Alumni Gari
Diana Odermatt, Sandra Kuster, Giulia Brunoldi, Alessandra Piceni, Laura Mariotti, Kerstin Gari, Sebastian Wild, Anna Simon
Dr. Kerstin Gari now in ZHAW School of Life Sciences and Facility Management, Wädenswil
IRON-SULPHUR PROTEINS AND GENOME STABILITY
The importance of genome stability
The blueprint of each cell – be it a single bacterium or part of a multi-cellular organism, such as man – is encoded in its genome. For a cell to remain functional throughout its life cycle, its genetic program has to remain accurate and, hence, its genome has to be protected from damage. Such damage can be the result of exogenous factors, such as UV light, or endogenous factors, such as the products of cellular metabolism. Moreover, the ability of a cell to duplicate its genome and divide into two identical daughter cells is a naturally complicated process that can introduce mistakes into the genetic information. To preclude these problems, cells have developed a multitude of mechanisms to protect their genomes: highly accurate factors for DNA replication, safeguard mechanisms that detect errors within the genome, and dedicated factors that can repair any possible damage within the DNA.
However, these protection mechanisms do not always work perfectly and if they fail, the consequences can be disastrous, cancer being the most renowned example. The genomes of cancer cells are intrinsically unstable, i.e. they display a high number of genetic mutations and chromosome rearrangements. As a consequence, their genetic program is altered, which can lead to uncontrolled cell growth and tumour development. Understanding in detail how genome stability is maintained is therefore an important step towards understanding cancer development.
Iron-sulphur proteins
Iron-sulphur (FeS) clusters are ancient and versatile cofactors that are found in a variety of proteins throughout all kingdoms of life. They are most commonly known for their role in electron transport in the mitochondrial respiratory chain, but they are also found in the active sites of a variety of enzymes. The most striking feature of FeS clusters is their ability to adopt redox potentials over a wide range, which is based on the fact that iron can switch between two oxidation states (+2 and +3).
A common motif is the so-called [4Fe-4S] cluster, in which four iron atoms and four sulphur atoms sit in the corners of a cube-like structure, which is incorporated into an FeS protein through coordinative binding by four cysteine residues (Figure 1).
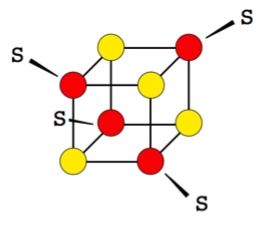
Figure 1: Schematic of a [4Fe-4S] cluster. Iron atoms are depicted in red, sulphur atoms in yellow. S denote cysteinyl ligands that coordinate the cluster.
Iron-sulphur proteins in DNA replication and repair
For a long time it appeared that nuclear FeS proteins with affinity for DNA are relatively rare. Given that upon FeS cluster oxidation, free iron atoms can generate reactive oxygen species and potentially damage DNA, this notion seemed rather intuitive.
However, recent evidence suggests that a considerable number of DNA replication and repair factors require binding to an FeS cluster for their function, amongst them the DNA helicases RTEL1, XPD, FANCJ and ChlR1, and proteins essential for DNA replication, such as DNA primase and DNA2.
With the discovery that all three replicative polymerases in yeast are FeS proteins, it has become clear that FeS clusters in DNA replication and repair proteins are the rule rather than the exception.
The maturation of nuclear FeS proteins takes place primarily in the cytoplasm, and is carried out by the cytoplasmic iron-sulphur assembly (CIA) machinery. We and others have shown that the HEAT-repeat protein MMS19, together with its interaction partner MIP18 , plays a key role in this process by physically linking the CIA machinery and FeS target proteins. By doing so, it facilitates the transfer of an FeS cluster to an FeS apoprotein and allows it to become a fully functional FeS holoprotein (Figure 2).
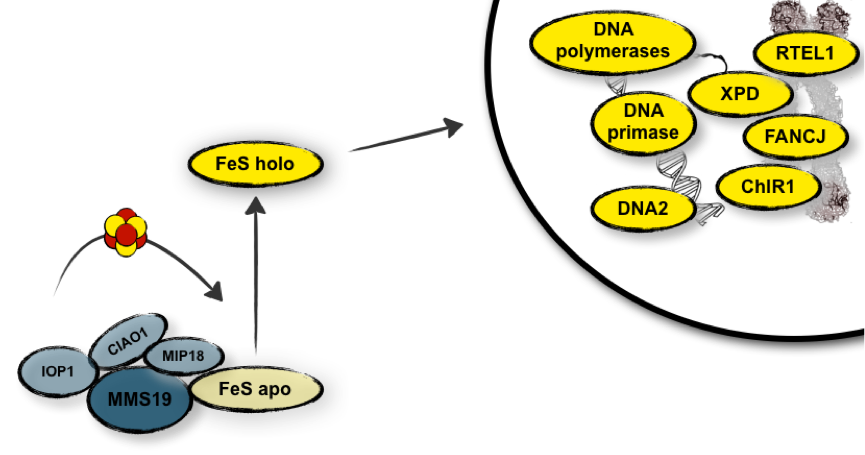
Figure 2: MMS19-MIP18 links cytoplasmic iron-sulphur cluster assembly to DNA metabolism.
In the absence of MMS19, when FeS protein maturation is impaired, the stability of FeS proteins and their assembly into functional complexes is affected. However, the actual function of FeS clusters in DNA replication and repair proteins has remained largely elusive to date.
Owing to their redox sensitivity, FeS clusters are particularly interesting and versatile cofactors that would be uniquely suited as regulatory cofactors that could e.g. sense oxidative stress conditions during DNA replication. One attractive possibility is that FeS clusters could serve as intrinsic stress sensors within DNA replication and repair proteins and directly modulate their biochemical activities to allow adaption to suboptimal conditions of DNA replication.
We are going to use a combination of techniques, such as biochemistry, molecular biology and structural studies, in order to address the function of FeS clusters in DNA replication and repair.