Non-Canonical Ubiquitination in the Regulation of DNA Damage Response and DNA Replication
Background
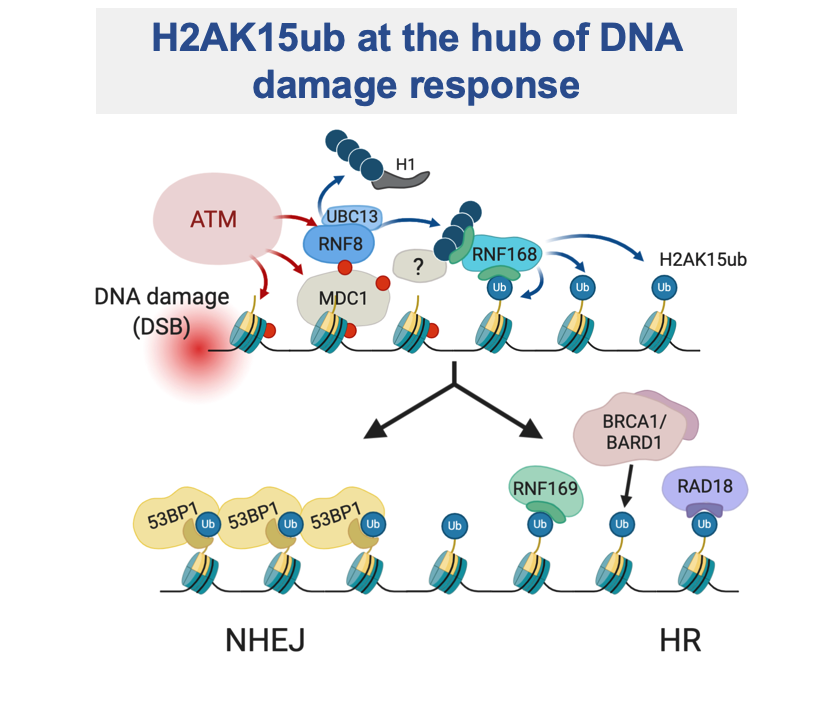
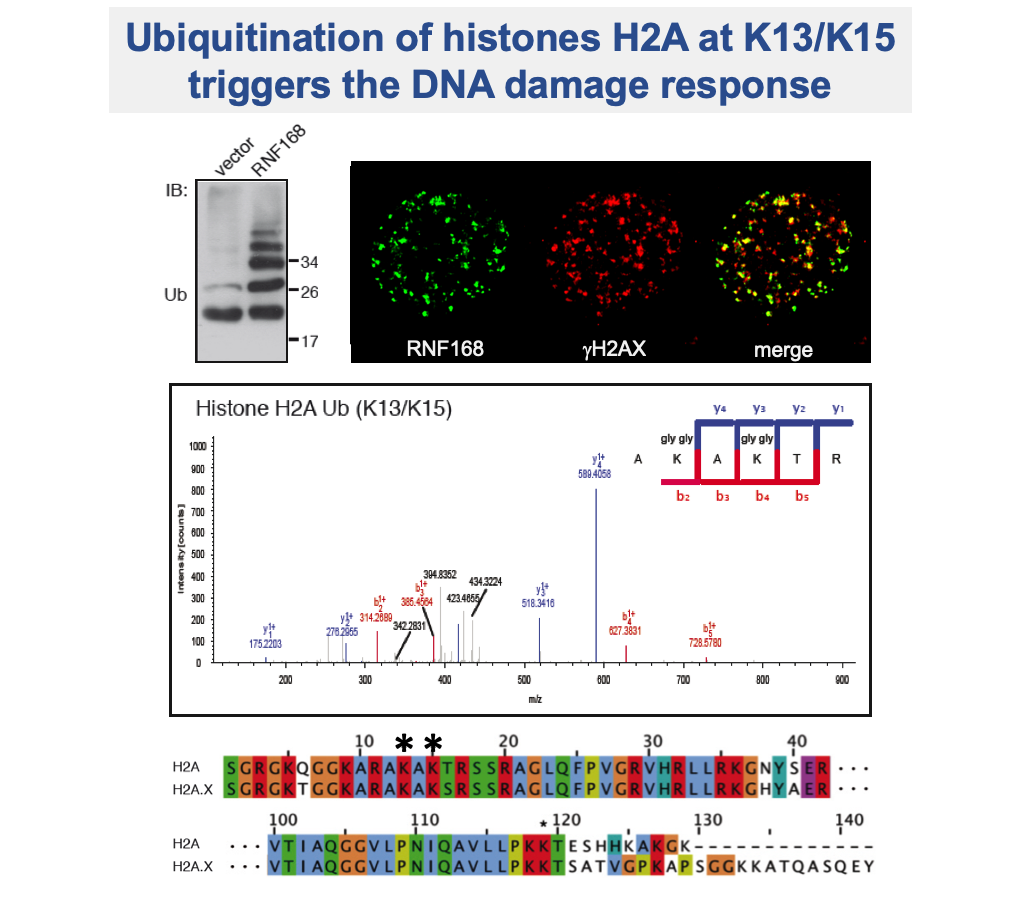
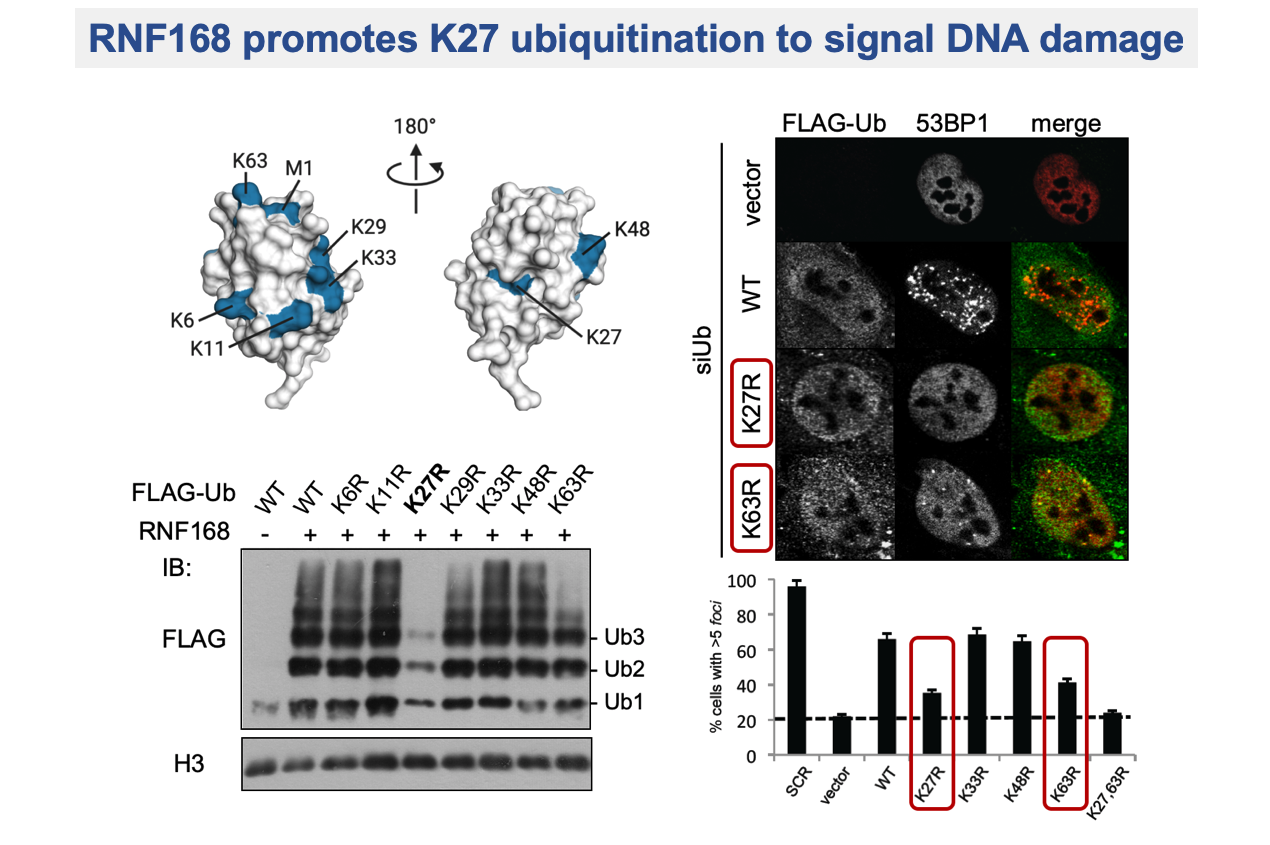
The non-degradative role of ubiquitination is essential to maintain genome stability by regulating the DNA damage response (DDR), DNA replication and cell cycle control. DNA lesions can lead, if unrepaired, to severe genomic alterations resulting in cell death or cancer formation. A variety of post-translational modifications occurs on damaged chromatin, triggering the recruitment of many proteins. A pivotal role is played by ubiquitination of histone H2As mediated by the ubiquitin ligase RNF168, which is essential to activate DNA repair factors. Prompted by the compelling question on how the specificity in DDR signalling is achieved, being H2As highly ubiquitinated at the C-terminal tail (H2AK119ub), we discovered that RNF168 exquisitely targets a novel site at N-terminal tail of H2A, yielding H2AK13ub and H2AK15ub. These novel histone marks have tremendous impact on the regulation of events occurring at damaged chromatin, being essential for promoting DNA repair. Further addressing the specificity of the RNF168-mediated signal, we found that K27-linked ubiquitination is required for RNF168 activity on histones and for triggering DDR signalling cascade.
Goal
Finding new determinants (post-translational modifications, writers, interacting/competing partners) of histone ubiquitination regulating chromatin remodelling in the DDR and DNA replication.
Ongoing and future work
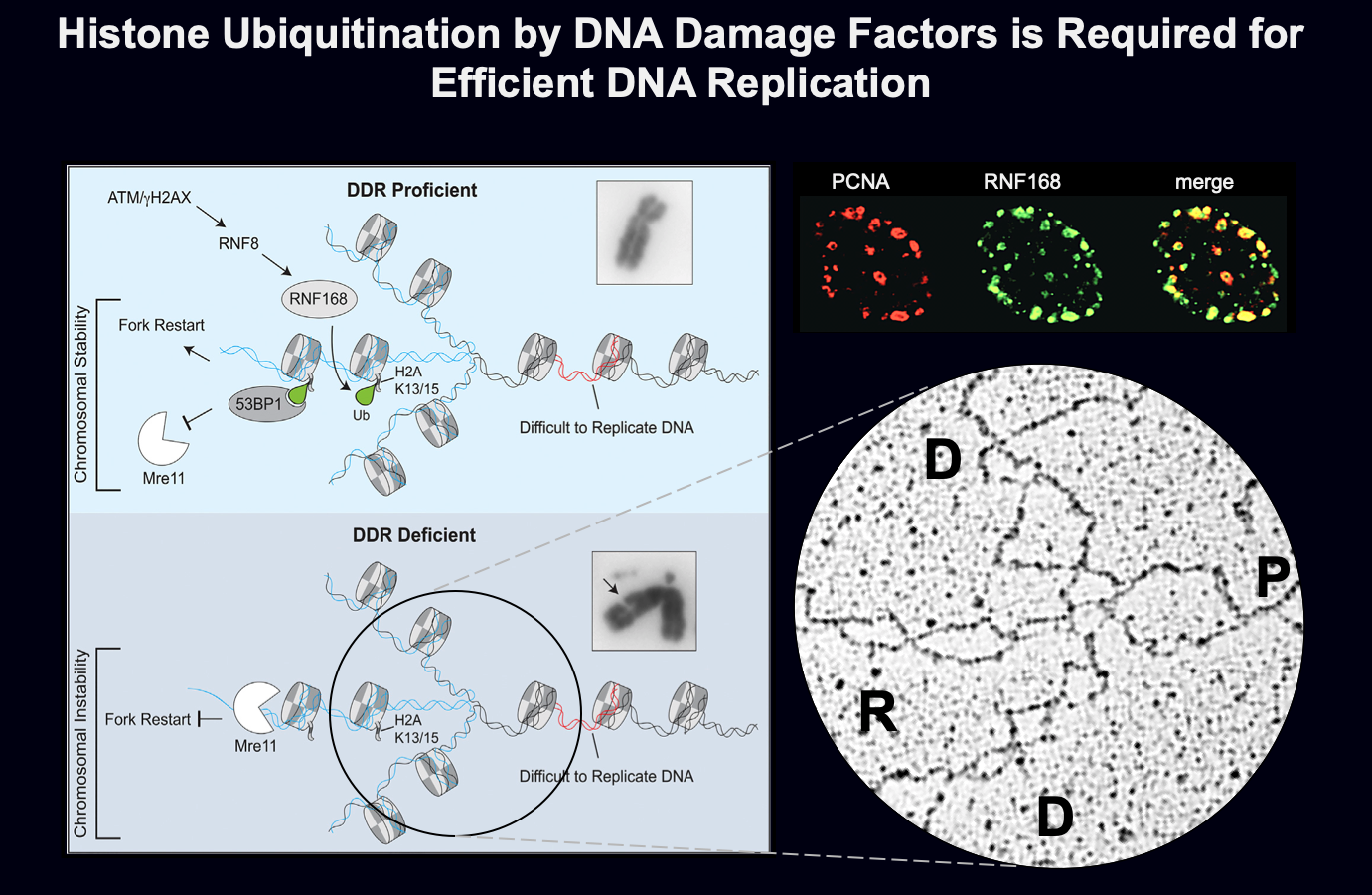
RNF168 and H2AK15ub in unperturbed DNA replication. We have recently found that the classical DDR factors, ATM, RNF8, RNF168 and 53BP1, are required for proper DNA replication in unperturbed conditions. Loss of RNF168 leads to reduced replication fork progression and to accumulation of replication intermediates (i.e. reversed forks), particularly evident at repetitive genomic sequences. Consistent with regular nucleosomal organization of reversed forks that we observed, RNF168 and its histone targets, H2AK13ub and H2AK15ub, are required for this replication function. We propose that double-stranded ends that form at reversed forks engage classical DDR factors, suggesting an alternative function of this pathway in preventing genome instability and human disease.
Role of DDR ubiquitin ligases in protection of stalled replication forks. We are currently expanding our understanding on the dynamics and the architecture of replication intermediates to investigate the role of ubiquitin ligases, including RNF168, in the processing and stability of DNA replication forks.
Selected publications
Mattiroli and Penengo L. Histone ubiquitination: an integrative signaling platform in genome stability. Trends in Genetics (2021).
Okonska A, Bühler S, Rao V, Ronner M, Blijlevens M, van der Meulen-Muileman IH, de Menezes RX, Wipplinger M, Oehl K, Smit EF, Weder W, Stahel RA, Penengo L, van Beusechem VW, Felley-Bosco E. Functional genomic screen in mesothelioma reveals that loss of function of BRCA1-associated-protein-1 induces chemoresistance to ribonucleotide reductase inhibition. Molecular Cancer Therapy (2019).
Schmid JA, Berti M, Walser F, Raso MC, Schmid F, Krietsch J, Stoy H, Zwicky K, Ursich S, Freire R, Lopes M, Penengo L. Histone ubiquitination by the DNA damage response is required for efficient DNA replication in unperturbed S-phase. Molecular Cell (2018).
Vujanovic M, Terraneo N, Zellweger R, Raso MR, Schmid JA, Taglialatela A, Holland CL, Zwicky K, Herrador R, Jacobs H, Cortez D, Ciccia A, Penengo L, Lopes M. Replication fork slowing and reversal upon genotoxic stress require PCNA polyubiquitination and ZRANB3 DNA translocase activity. Molecular Cell (2017).
Balbo Pogliano C, Gatti M, Rüthemann P, Garajovà Z, Penengo L, Naegeli H. ASH1L histone methyltransferase stimulates global-genome nucleotide excision repair. Nature Communications, (2017).
Graf U, Casanova EA, Wyck S, Dalcher D, Gatti M, Vollenweider E, Okoniewski MJ, Weber F, Patel SS, Schmid M, Li J, Sharif J, Wanner G, Koseki H, Wong J, Pelczar P, Penengo L, Santoro R, Cinelli P. Pramel7 mediates ground state pluripotency through proteasomal-epigenetic combined pathways. Nature Cell Biology (2017).
Parrotta R, Okonska A, Ronner M, Weder W, Stahel R, Penengo L, Felley-Bosco E. A novel BRCA1-associated protein-1 isoform affects response of mesothelioma cells to drugs impairing BRCA1-mediated DNA repair. Journal of thoracic oncology (2017).
Gatti M, Pinato S, Maiolica A, Rocchio F, Prato MG, Aebersold R, Penengo L. RNF168 promotes non-canonical K27 ubiquitination to signal DNA damage. Cell Reports (2015).
Gatti M, Pinato S, Maspero E, Soffientini P, Polo S, Penengo L: A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle (2012).
Pinato S, Gatti M, Scandiuzzi C, Confalonieri S, Penengo L: UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Molecular Cell Biology (2011).
Pinato S, Scandiuzzi C, Arnaudo N, Citterio E, Gaudino G, Penengo L: RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Molecular Biology (2009).