Causes of transcription-replication conflicts
Background
To ensure accurate duplication of the genome, replication forks must overcome numerous obstacles including transcription complexes moving along the same DNA template. Transcription-replication encounters can occur in either co-directional or head-on orientation, with the latter scenario having a strong deleterious effect on replication fork progression and genomic integrity. Recent studies have shown that head-on transcription-replication conflicts (TRCs) promote the formation of co-transcriptional R-loops that represent a strong block to replication fork progression. R-loops are three-stranded structures generated by invasion of the nascent transcript into the underwound DNA duplex behind the transcription complex, leading to the formation of an RNA:DNA hybrid between the transcript and the template DNA strand. R-loop formation is favored in regions with a high G density in the non-transcribed strand, and evidence suggests that formation of G-quadruplex structures in this strand stabilizes the R-loop structure and promotes its extension. Importantly, R-loops but not normal transcription complexes induce DNA breakage during head-on TRCs, thus implicating R-loops as the major cause of replication stalling induced by head-on transcription. Accumulating evidence suggest that the frequency of transcription-dependent fork breakage is elevated in cells overexpressing oncogenes that induce firing of intragenic replication origins (e.g. Cyclin E, c-Myc) or increase global transcription activity (e.g. HRasV12). However, the question remains whether or not this oncogene-induced replication stress is caused by R-loops.
Goal
The goal of our research is to provide a comprehensive understanding of the factors and underlying molecular mechanisms that can induce R-loop-mediated replication stress.
Ongoing projects
Role of co-transcriptional R-loops in oncogene-induced replication stress.
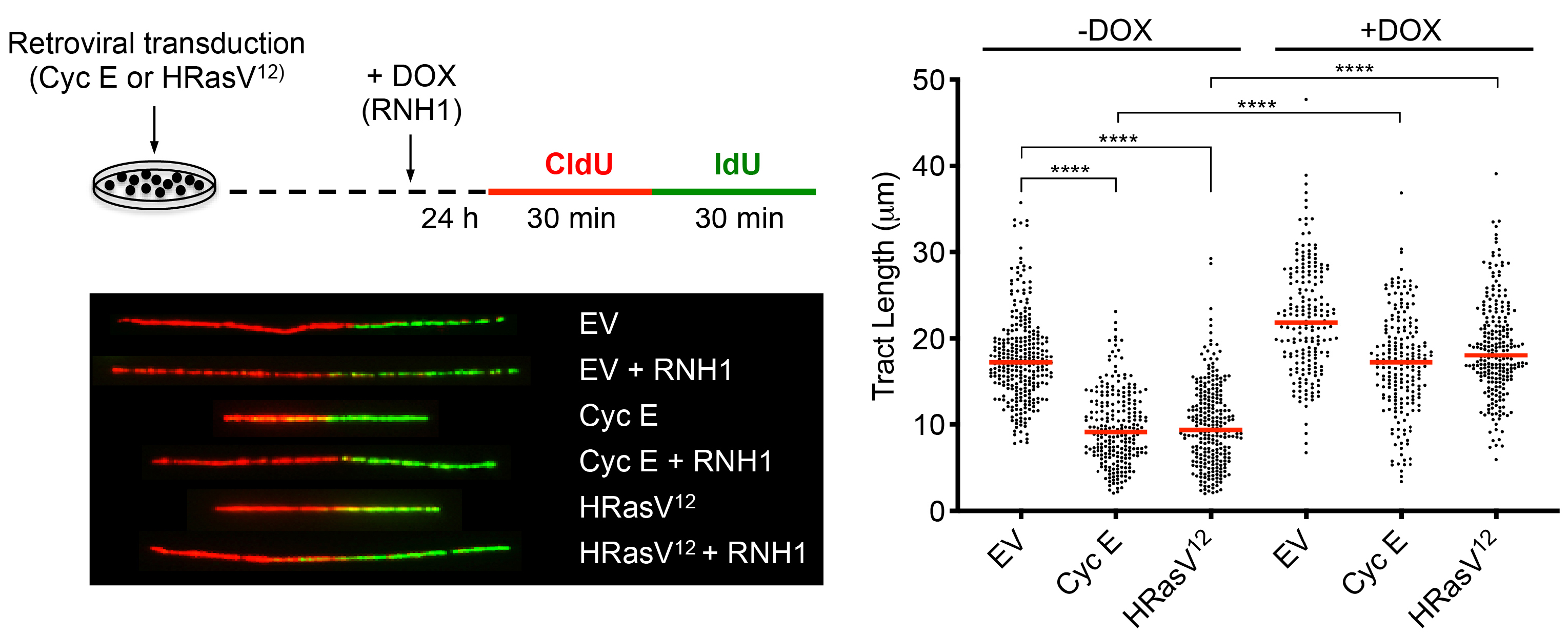
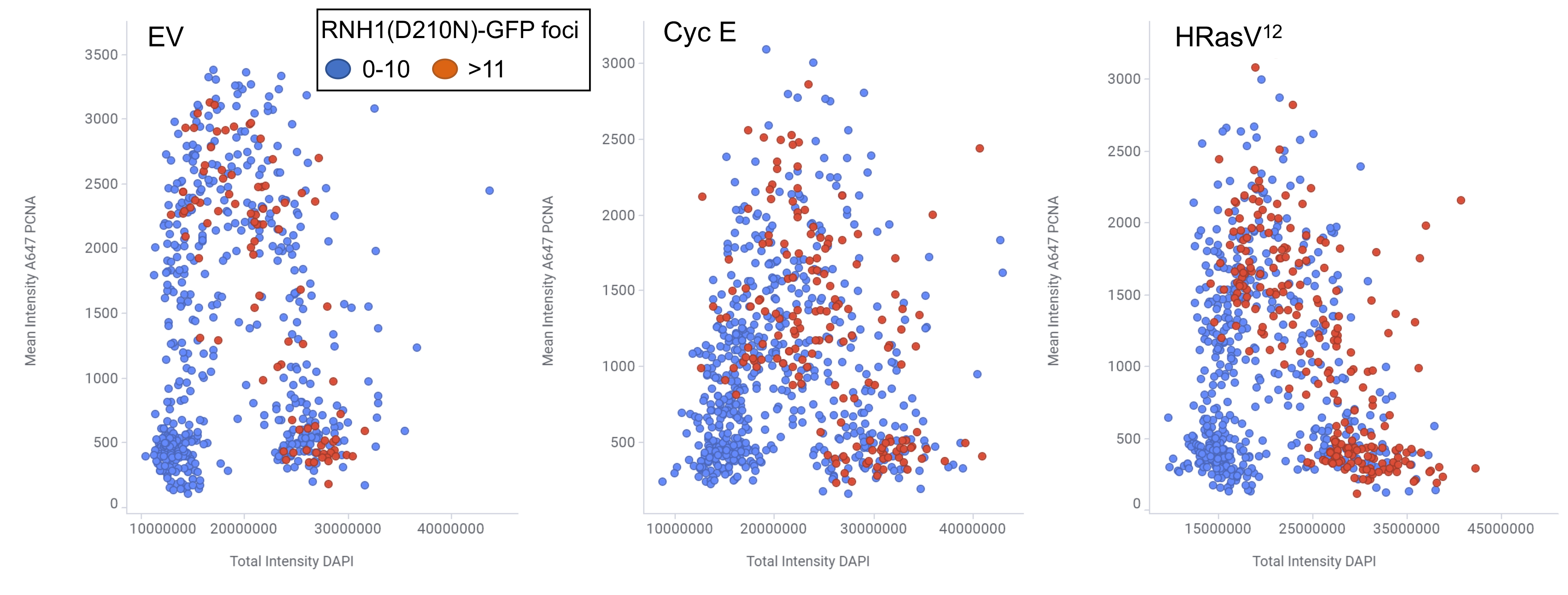
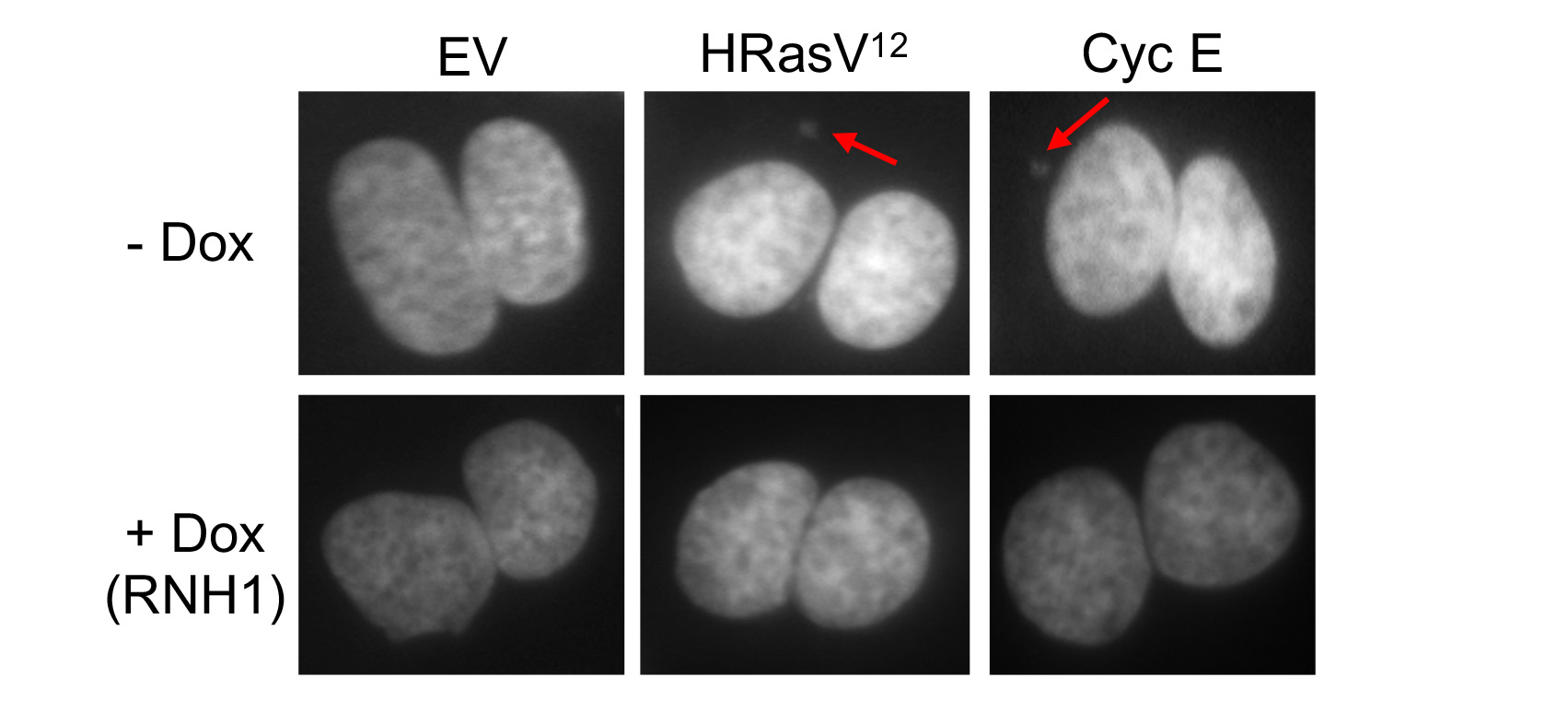
We have been testing the hypothesis that oncogene-induced replication stress results from R-loop-mediated TRCs. Our work has shown that replication fork slowing and DNA damage (micronuclei formation) induced by the Cyclin E and HRasV12 oncogenes can be completely rescued by overexpression of RNase H1, which eliminates R-loops by cleaving the RNA strand in the RNA:DNA hybrid. Moreover, overexpression of these oncogenes induced the accumulation of nuclear foci of the catalytically-inactive RNase H1, indicative of R-loop formation. We are now using chromatin immunoprecipitation (ChIP) combined with next generation sequencing (ChIP-Seq) to identify the sites of R-loop formation (ChIP of RNaseH1(D210N)-GFP bound hybrids) and replisome stalling (ChIP of DNA pol epsilon) in cells overexpressing the HRasV12 and Cyclin E oncogenes, and will compare the resulting data with existing databases of mutation hotspots in cancer samples. We also collaborate with the laboratory of Prof. Anne Müller to investigate the molecular mechanism of R-loop-dependent replication stress detected upon activation of the oncogenic NF-kB signaling pathway by Helicobacter pylori infection.
Oxidative stress and R-loop-mediated replication fork stalling.
A recent study has shown that reactive oxygen species (ROS) generated upon treatment of cells with the ribonucleotide reductase inhibitor hydroxyurea (HU) compromise binding of the fork accelerator TIMELESS to the replisome, leading to replication stress. As HU stimulates TRCs at genomic loci termed early replication fragile sites, we seek to determine whether replication fork stalling induced by oxidative stress is a consequence of R-loop-mediated TRCs. Our hypothesis is that R-loops are formed if the transcription complex collides with a functionally impaired replisome.
DNA lesion-arrested transcription complexes as source of R-loop-mediated replication stress.
Bulky DNA adducts and inter-strand crosslinks can halt the progression of transcription and replication machineries. While there are mechanisms for the replisome to bypass such DNA lesions, RNA polymerase does not have such capability and stalls after encounter of the lesion generating a roadblock for oncoming replication fork. RNA polymerase stalling may lead to the formation of an R-loop. We will examine whether R-loop formation underlies replication fork stalling in cells exposed to agents that induce transcription-blocking lesions including mitomycin C, UV-C light or cisplatin.
Selected publications
Bauer M, Nascakova Z, Mihai AI, Cheng PF, Levesque MP, Lampart S, Hurwitz R, Pfannkuch L, Dobrovolna J, Jacobs M, Bartfeld S, Dohlman A, Shen X, Gall AA, Salama NR, Töpfer A, Weber A, Meyer TF, Janscak P*, Müller A.* (2020) The ALPK1/TIFA/NF-κB axis links a bacterial carcinogen to R-loop-induced replication stress. Nat. Commun. 11:5117.
*corresponding authors