Resolution of transcription-replication conflicts
Background
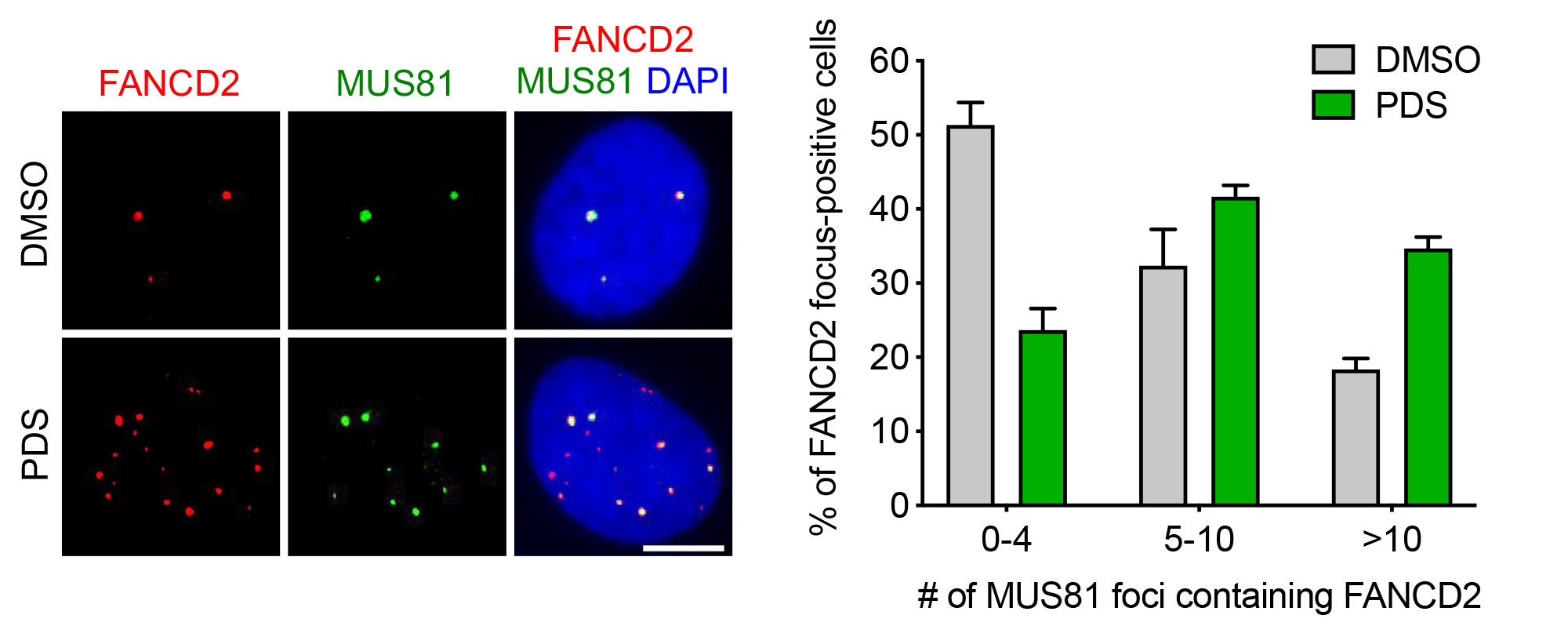
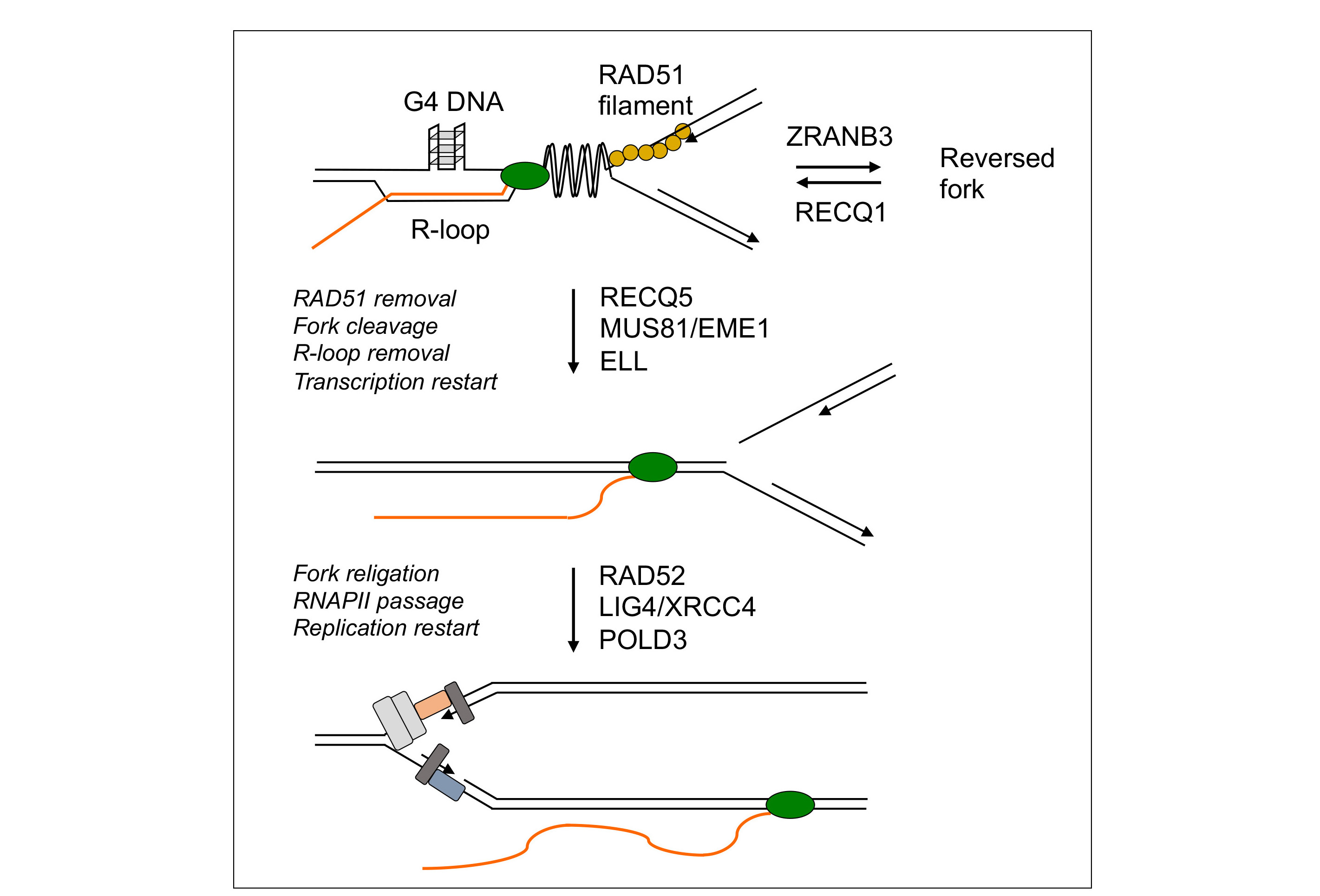
DNA transactions occurring at sites of TRCs to maintain genomic stability are not well understood. We have studied those transactions upon induction of TRCs by drugs that promote R-loop formation (camptothecin, pyridostatin, aphidicolin). Our study revealed that replication fork stalling by co-transcriptional R-loops is an active process involving replication fork reversal mediated by RAD51 recombinase and the DNA translocase ZRANB3. Moreover, we have discovered that RECQ1-mediated reverse branch migration on the resulting four-way DNA structure triggers the restart of semiconservative DNA replication, which requires RECQ5 DNA helicase, MUS81-EME1 endonuclease, the DNA ligase IV(LIG4)-XRCC4 complex, the non-catalytic subunit of DNA polymerase δ, POLD3, the transcription elongation factor ELL and active RNA synthesis. Detailed analysis of the underlying molecular mechanism suggested a model for TRC resolution where a fork cleavage-religation cycle catalyzed by MUS81-EME1 and LIG4-XRCC4 relieves the torsional stress in the DNA template generated by TRC, allowing bypass of the transcription complex across the replication-stalling site and subsequent replication restart. We have also found that RECQ5 helicase removes RAD51 filament from the fork junction to prevent a new round of fork reversal and to facilitate fork cleavage by MUS81-EME1.
Goal
The aim of this project is to gain further insights into the molecular mechanisms underlying MUS81-dependent restart of R-loop-stalled replication forks in human cells.
Ongoing work
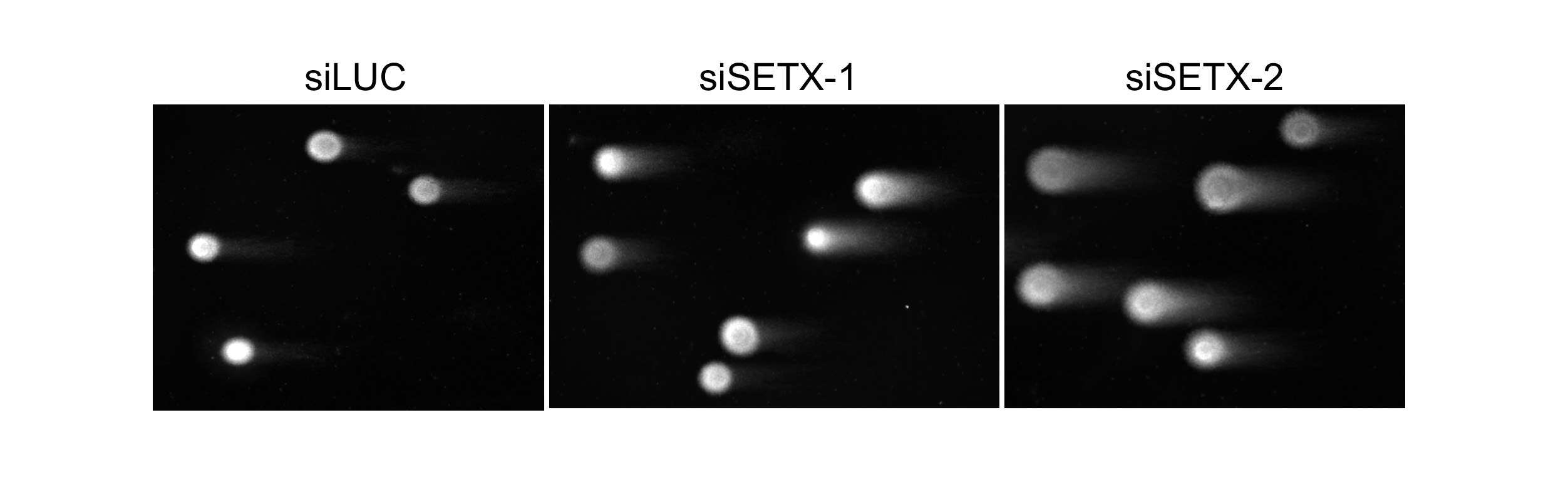
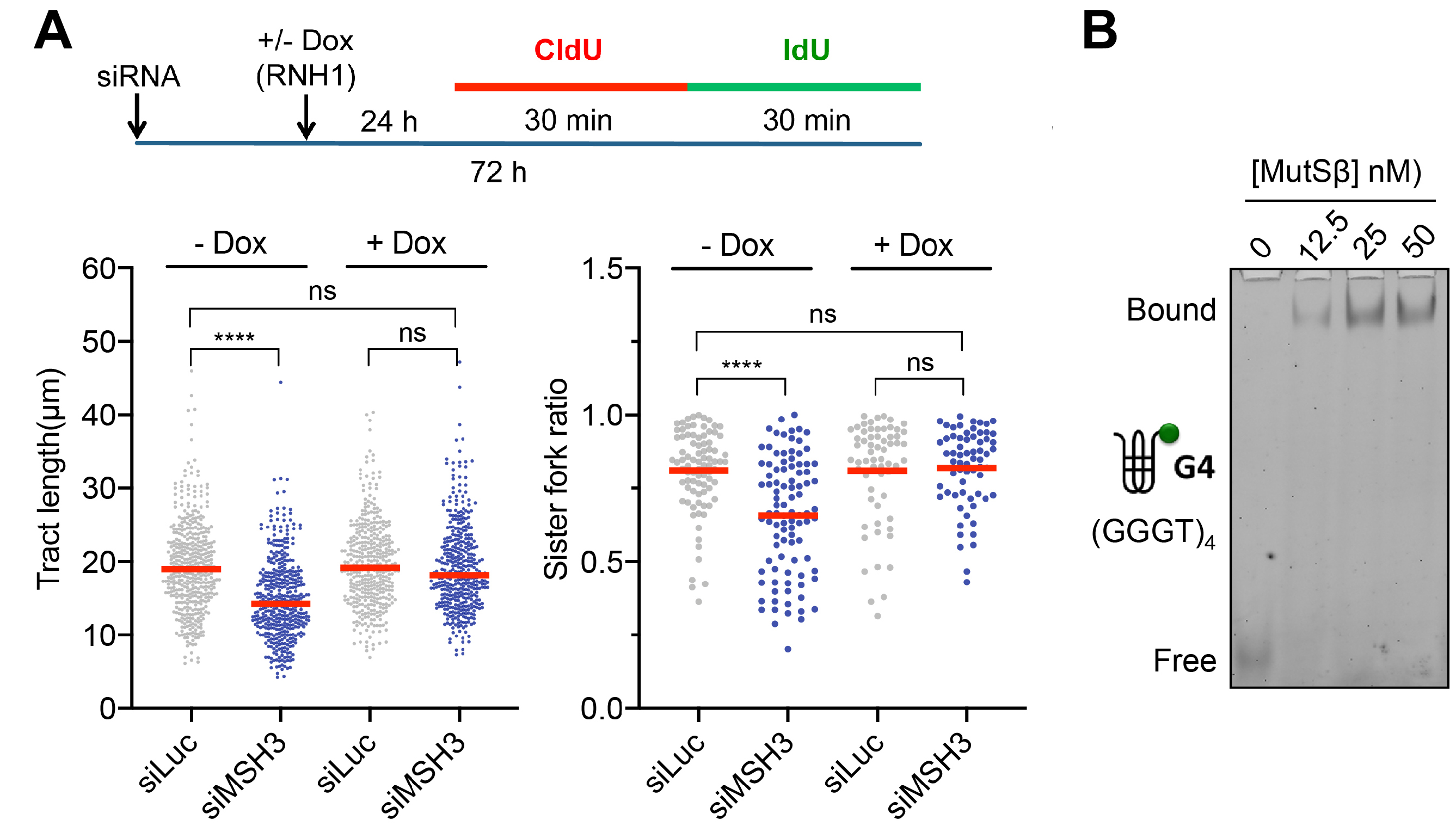
Our current research focuses on the identification of the proteins and underlying mechanisms involved in the elimination R-loops and associated G-quadruplex structures at TRC sites, which is required to allow the passage of the reactivated replication fork. Based on our preliminary experiments and results of previous studies, we focus on the RNA/DNA helicase Senataxin and the DNA loop-binding factor MutSβ. Furthermore, we apply complementary mass spectrometry-based proteomics approaches, including APEX-based proximity labeling and chromatin affinity precipitation methods, coupled with a functional RNA interference screening to identify new factors involved in these processes.
Selected publications
Chappidi N, Nascakova Z, Boleslavska B, Zellweger R, Isik E, Andrs M, Menon S, Dobrovolna J, Balbo Pogliano C, Matos J, Porro A, Lopes M, Janscak P (2020) Fork cleavage-religation cycle and active transcription mediate replication restart after fork stalling at co-transcriptional R-loops. Mol. Cell 77(3), 528-541.
Di Marco S, Hasanova Z, Kanagaraj R, Chappidi N, Altmannova V, Menon S, Sedlackova H, Langhoff J, Surendranath K, Hühn D, Bhowmick R, Marini V, Ferrari S, Hickson ID, Krejci L, Janscak P. (2017) RECQ5 Helicase Cooperates with MUS81 Endonuclease in Processing Stalled Replication Forks at Common Fragile Sites during Mitosis. Mol. Cell 66(5), 658-671.
Urban V, Dobrovolna J, Hühn D, Fryzelkova J, Bartek J, Janscak P. (2016) RECQ5 helicase promotes resolution of conflicts between replication and transcription in human cells. J. Cell Biol. 214(4), 401-15.