Nuclear dynamics in response to replication stress
Background
Interference with DNA replication is a prominent clinical strategy to counteract uncontrolled cancer cell-proliferation, with the rationale that the majority of non-cancerous cells is quiescent and thus remains largely unaffected. A major limitation for effective cancer therapy however is the limited knowledge on the mechanism of action of most chemotherapeutic regimens. Understanding how the replication apparatus is specifically challenged by each treatment and which factors assist the cellular response to these perturbations is instrumental to any strategy aiming to improve cancer chemotherapy and to limit chemoresistance, an outcome frequently observed in cancer patients.
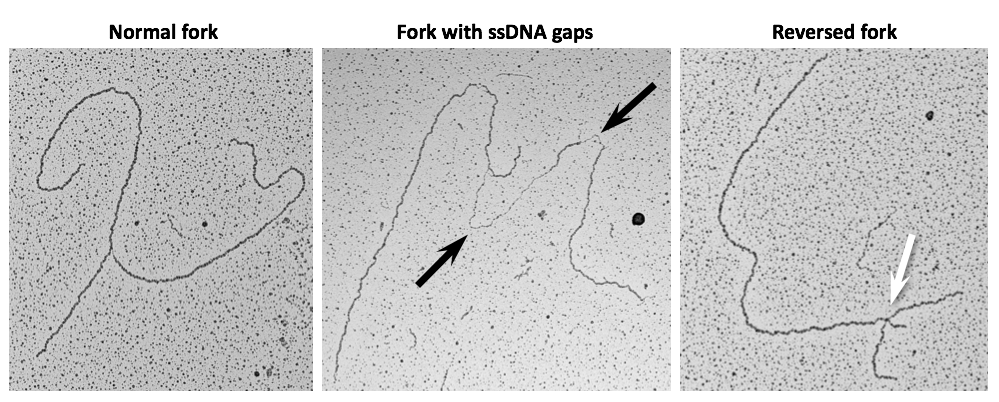
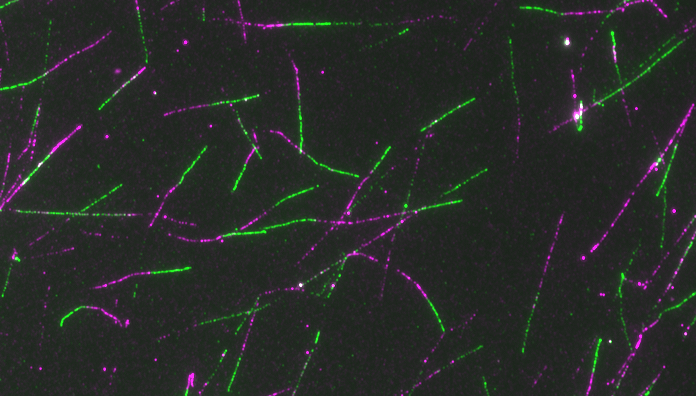
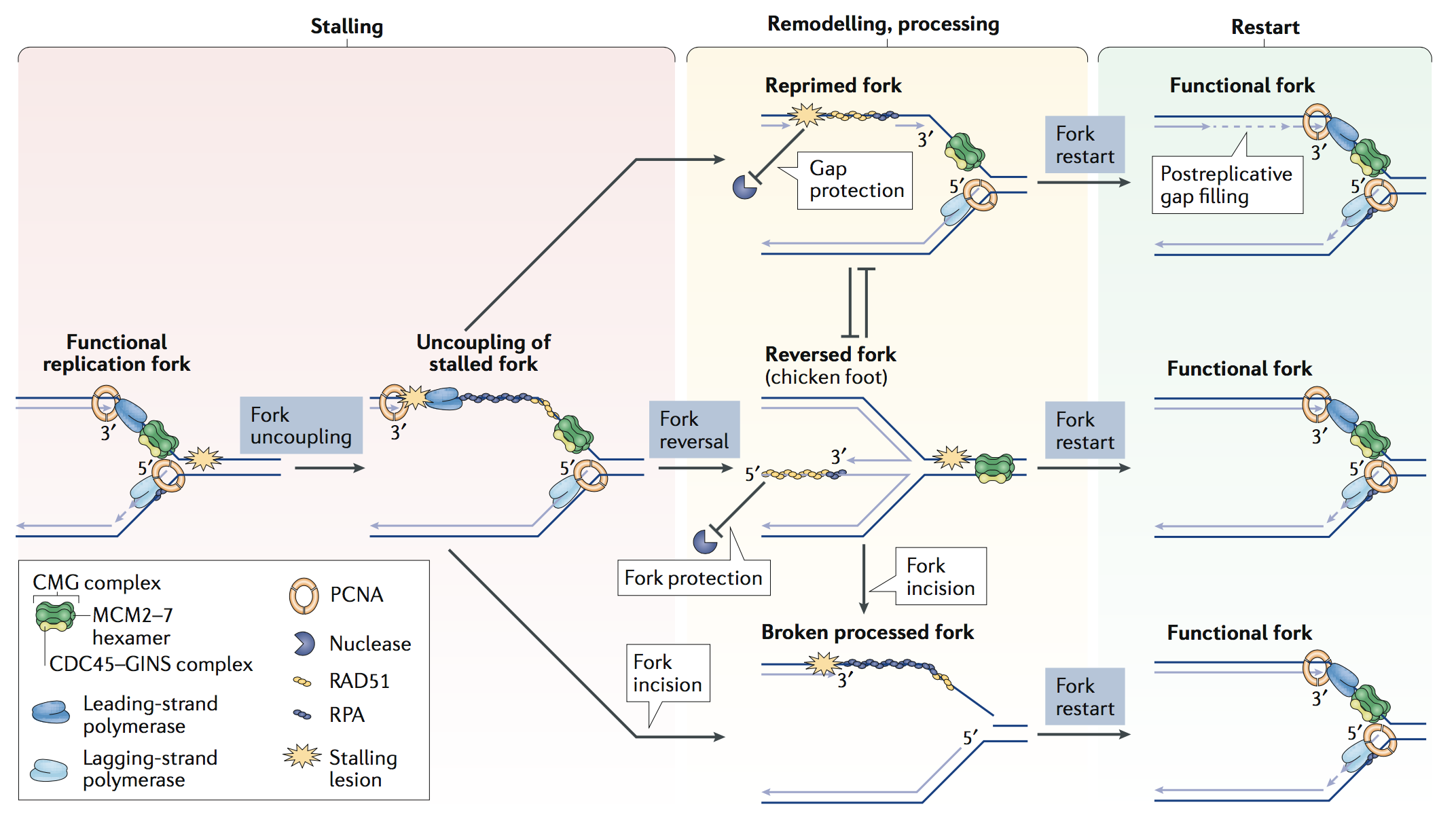
We successfully refined specialized single-molecule approaches on replication intermediates and pioneered their use on cancer cell lines to uncover molecular responses to drug-induced replication stress. Thereby we have provided the first direct visualization of replication fork remodelling into four-way junctions - namely fork reversal - in human cells, as a global, transient, evolutionarily conserved response to various perturbations of the replication process (Ray Chaudhuri et al., Nat Str Mol Biol 2012; Zellweger et al., J Cell Biol 2015).
These studies allowed refining long-standing models on the mechanism of action of commonly used anticancer drugs. Recently, our lab and others have reported that fork remodelling and restart in face of obstacles are assisted by key components of nuclear architecture. How nuclear dynamics helps integrating local transactions into a global response to replication stress within the intact nucleus has remained elusive and is a fascinating area of future research.
Goal
We aim to assess the role of nuclear architectural components (e.g. nuclear F-Actin, Lamin A and Cohesin) in replication fork plasticity, and test how their structural reorganization assists challenged replication domains. While continuing to exploit our powerful single-molecule approaches, we are currently expanding our portfolio of genomic and advanced light micrsocopy approaches to investigate at high resolution how the replication stress response is orchestrated spatiotemporally within the nucleus.
Ongoing and future work
We are currently studying how 3D genome interactions – known to orchestrate the replication program in unperturbed conditions – may impact the response to local and global RS. Moreover, we are exploiting new probes and super-resolution imaging to visualize how replication factories react to genotoxic stress, recruiting specialised fork plasticity factors and taking contact with key components of nuclear architecture.
Combining single-molecule and single-cell assays, super-resolution imaging and state-of-the-art genomics, we aim to decipher the RS response at unprecedented resolution and from a new perspective, revealing the dynamic rearrangement of nuclear architectural components that mediate the rapid response to clinically relevant replication interference.
Recent selected publications
R. Zellweger and M. Lopes (2018). Dynamic Architecture of Eukaryotic DNA Replication Forks In Vivo, Visualized by Electron Microscopy. In "Genome Instability". Humana Press, ed. G. Brown and M. Muzi Falconi. Methods in Molecular Biology, 1672:261-294
K. Mutreja, J. Krietsch, J. Hess, S. Ursich, M. Berti, F.K. Roessler, R. Zellweger, M. Patra, G. Gasser and M. Lopes (2018). ATR-Mediated Global Fork Slowing and Reversal Assist Fork Traverse and Prevent Chromosomal Breakage at DNA Interstrand Cross-Links. Cell Reports, 24(10):2629-2642.e5.
M. Berti, D. Cortez and M. Lopes (2020). The plasticity of DNA replication forks in response to clinically-relevant genotoxic stress. Nature Reviews Mol Cell Biol.
Palumbieri, Maria Dilia; Merigliano, Chiara; González-Acosta, Daniel; Kuster, Danina; Krietsch, Jana; Stoy, Henriette; von Känel, Thomas; Ulferts, Svenja; Welter, Bettina; Frey, Joël; Doerdelmann, Cyril; Sanchi, Andrea; Grosse, Robert; Chiolo, Irene; Lopes, Massimo (2023). Nuclear actin polymerization rapidly mediates replication fork remodeling upon stress by limiting PrimPol activity. Nature Communications, 14(1):7819.
Ulferts, Svenja; Lopes, Massimo; Miyamoto, Kei; Grosse, Robert (2024).Nuclear actin dynamics and functions at a glance. Journal of Cell Science, 137(6):jcs261630.
González-Acosta, Daniel; Lopes, Massimo (2024).DNA replication and replication stress response in the context of nuclear architecture. Chromosoma, 133(1):57-75.
Veronica Cherdyntseva, Joanna Paulson, Selin Adakli, Jean-Philippe Gagné, Moses Aouami, Patricia Ubieto-Capella, Daniel González-Acosta, Collin Bakker, Guy G. Poirier, Nitika Taneja, Massimo Lopes (2025). Nucleoplasmic Lamin A/C controls replication fork restart upon stress by modulating local H3K9me3 and ADP-ribosylation levels. bioRxiv, doi: https://doi.org/10.1101/2025.01.18.633737